Mastering Custom SageMaker Deployment: A Comprehensive Guide
A deep dive into the intricacies of deploying custom models to Amazon SageMaker

import pandas as pd
import numpy as np
import seaborn as sns
import torch
import torch.nn as nn
import torch.nn.functional as F
from torch.utils.data import DataLoader
from torch.utils.data import TensorDataset
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score, confusion_matrix, classification_report
from sklearn.preprocessing import StandardScaler
# set default figure size
plt.rcParams['figure.figsize'] = (15, 7.0)
heart_data = '../input/heart-attack-analysis-prediction-dataset/heart.csv'
heart_df = pd.read_csv(heart_data)
heart_df.head()
| age | sex | cp | trtbps | chol | fbs | restecg | thalachh | exng | oldpeak | slp | caa | thall | output | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 63 | 1 | 3 | 145 | 233 | 1 | 0 | 150 | 0 | 2.3 | 0 | 0 | 1 | 1 |
| 1 | 37 | 1 | 2 | 130 | 250 | 0 | 1 | 187 | 0 | 3.5 | 0 | 0 | 2 | 1 |
| 2 | 41 | 0 | 1 | 130 | 204 | 0 | 0 | 172 | 0 | 1.4 | 2 | 0 | 2 | 1 |
| 3 | 56 | 1 | 1 | 120 | 236 | 0 | 1 | 178 | 0 | 0.8 | 2 | 0 | 2 | 1 |
| 4 | 57 | 0 | 0 | 120 | 354 | 0 | 1 | 163 | 1 | 0.6 | 2 | 0 | 2 | 1 |
# describe the data
heart_df.describe()
| age | sex | cp | trtbps | chol | fbs | restecg | thalachh | exng | oldpeak | slp | caa | thall | output | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 303.000000 | 303.000000 | 303.000000 | 303.000000 | 303.000000 | 303.000000 | 303.000000 | 303.000000 | 303.000000 | 303.000000 | 303.000000 | 303.000000 | 303.000000 | 303.000000 |
| mean | 54.366337 | 0.683168 | 0.966997 | 131.623762 | 246.264026 | 0.148515 | 0.528053 | 149.646865 | 0.326733 | 1.039604 | 1.399340 | 0.729373 | 2.313531 | 0.544554 |
| std | 9.082101 | 0.466011 | 1.032052 | 17.538143 | 51.830751 | 0.356198 | 0.525860 | 22.905161 | 0.469794 | 1.161075 | 0.616226 | 1.022606 | 0.612277 | 0.498835 |
| min | 29.000000 | 0.000000 | 0.000000 | 94.000000 | 126.000000 | 0.000000 | 0.000000 | 71.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 47.500000 | 0.000000 | 0.000000 | 120.000000 | 211.000000 | 0.000000 | 0.000000 | 133.500000 | 0.000000 | 0.000000 | 1.000000 | 0.000000 | 2.000000 | 0.000000 |
| 50% | 55.000000 | 1.000000 | 1.000000 | 130.000000 | 240.000000 | 0.000000 | 1.000000 | 153.000000 | 0.000000 | 0.800000 | 1.000000 | 0.000000 | 2.000000 | 1.000000 |
| 75% | 61.000000 | 1.000000 | 2.000000 | 140.000000 | 274.500000 | 0.000000 | 1.000000 | 166.000000 | 1.000000 | 1.600000 | 2.000000 | 1.000000 | 3.000000 | 1.000000 |
| max | 77.000000 | 1.000000 | 3.000000 | 200.000000 | 564.000000 | 1.000000 | 2.000000 | 202.000000 | 1.000000 | 6.200000 | 2.000000 | 4.000000 | 3.000000 | 1.000000 |
# checking data types
heart_df.dtypes
age int64
sex int64
cp int64
trtbps int64
chol int64
fbs int64
restecg int64
thalachh int64
exng int64
oldpeak float64
slp int64
caa int64
thall int64
output int64
dtype: object
# drop duplicates if any
heart_df.drop_duplicates()
# check missing valus
heart_df.isna().sum()
age 0
sex 0
cp 0
trtbps 0
chol 0
fbs 0
restecg 0
thalachh 0
exng 0
oldpeak 0
slp 0
caa 0
thall 0
output 0
dtype: int64
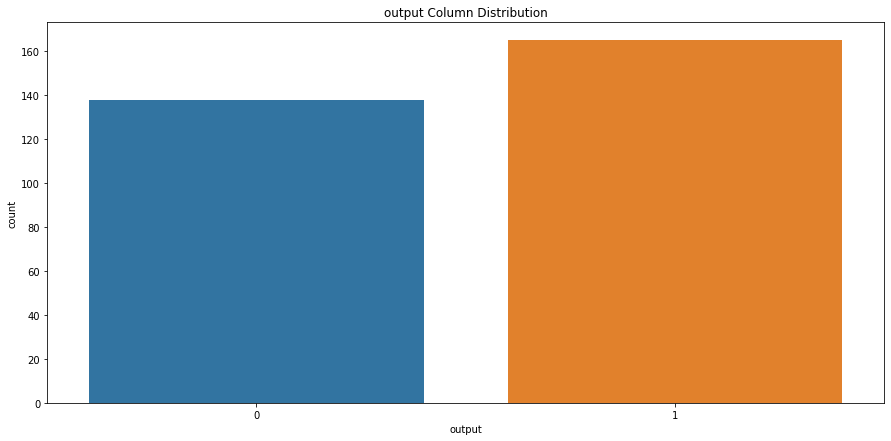
# check output column class distribution
sns.countplot(x='output', data=heart_df).set_title("output Column Distribution")
Text(0.5, 1.0, 'output Column Distribution')
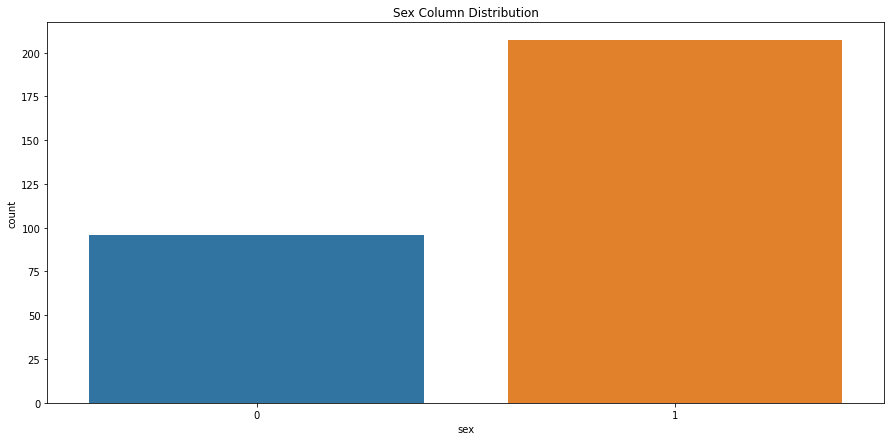
# check sex column class distribution
sns.countplot(x='sex', data=heart_df).set_title("Sex Column Distribution")
Text(0.5, 1.0, 'Sex Column Distribution')
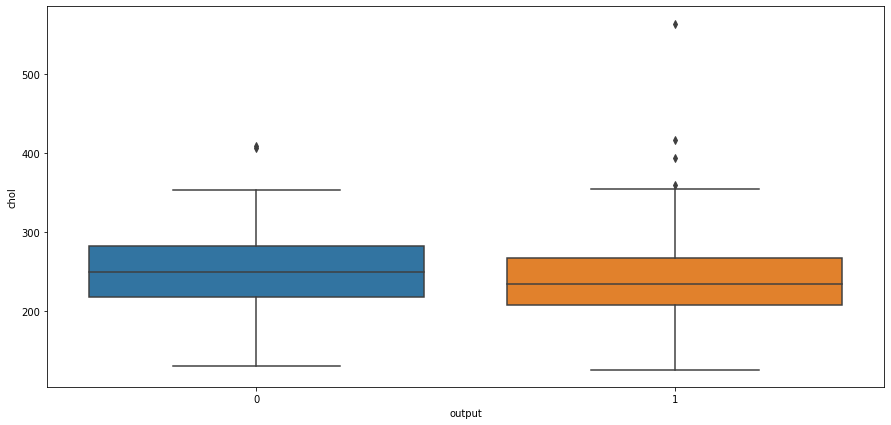
# box plot for output and cholestrol level
sns.boxplot(x="output",y="chol",data=heart_df)
<AxesSubplot:xlabel='output', ylabel='chol'>
# box plot for output and cholestrol level
sns.boxplot(x="output",y="thalachh",data=heart_df)
<AxesSubplot:xlabel='output', ylabel='thalachh'>
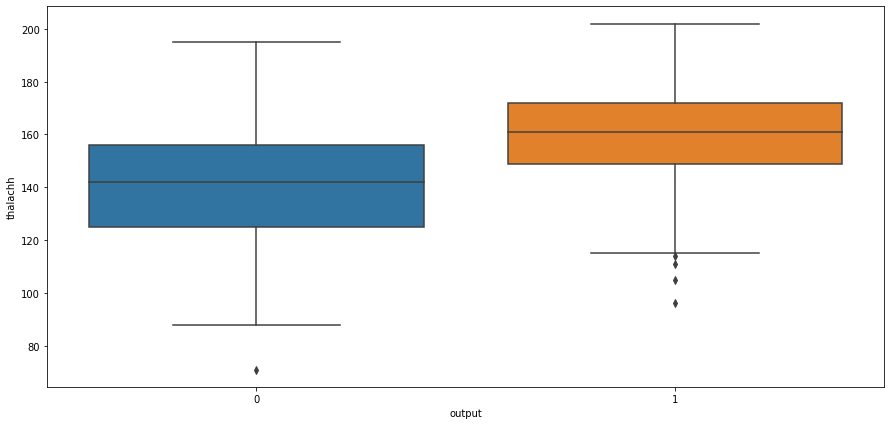
# box plot for output and cholestrol level
sns.boxplot(x="output",y="oldpeak",data=heart_df)
<AxesSubplot:xlabel='output', ylabel='oldpeak'>
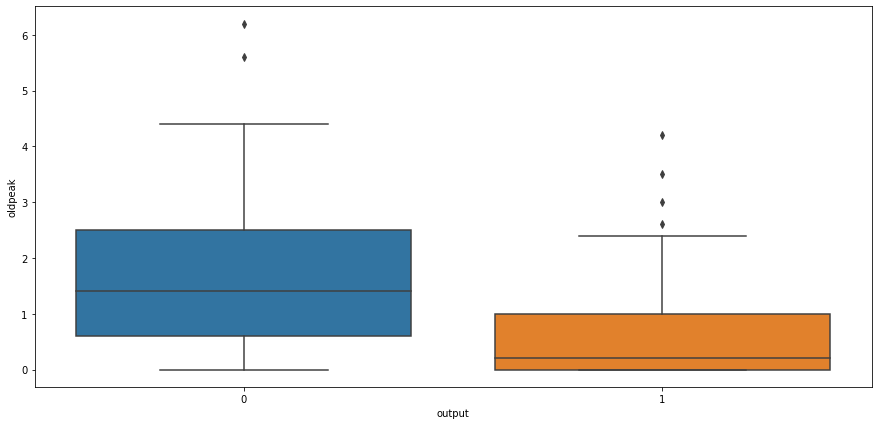
# box plot for output and cholestrol level
sns.boxplot(x="output",y="age",data=heart_df)
<AxesSubplot:xlabel='output', ylabel='age'>
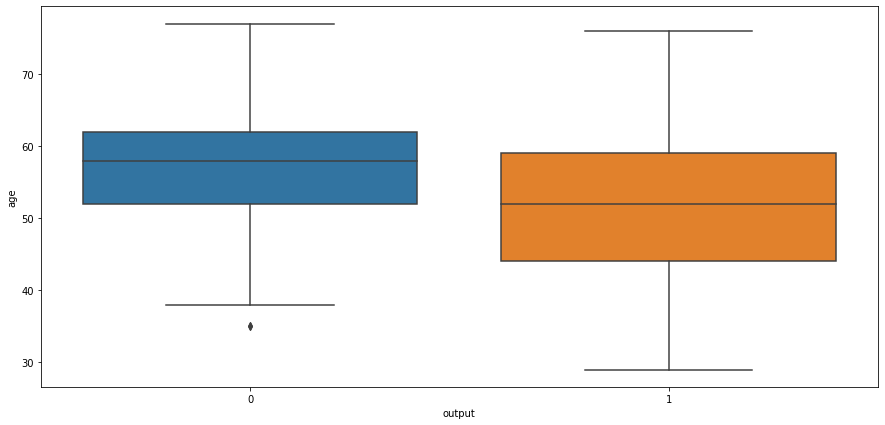
ax = sns.countplot(x='age', data=heart_df)
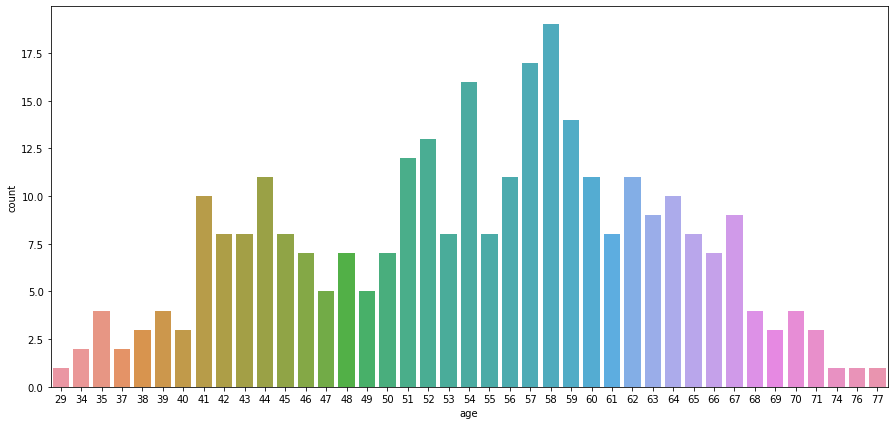
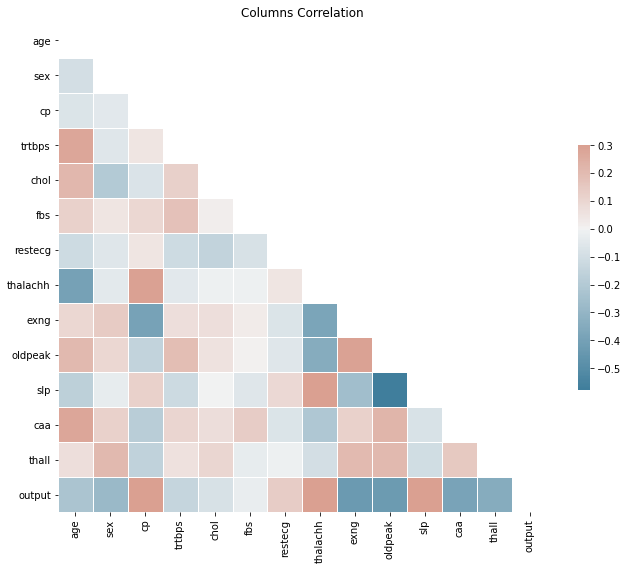
# check correlation
corr = heart_df.corr()
# Generate a mask for the upper triangle
mask = np.triu(np.ones_like(corr, dtype=bool))
# Set up the matplotlib figure
f, ax = plt.subplots(figsize=(11, 9))
# Generate a custom diverging colormap
cmap = sns.diverging_palette(230, 20, as_cmap=True)
# Draw the heatmap with the mask and correct aspect ratio
sns.heatmap(corr, mask=mask, cmap=cmap, vmax=.3, center=0,
square=True, linewidths=.5, cbar_kws={"shrink": .5}).set_title("Columns Correlation")
Text(0.5, 1.0, 'Columns Correlation')
# split data for training
y = heart_df.output.to_numpy()
X = heart_df.drop('output', axis=1).to_numpy()
# scale X values
scaler = StandardScaler()
X = scaler.fit_transform(X)
# split data while keeping output class distribution consistent
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, stratify=y)
# convert data to pytorch tensors
def df_to_tensor(df):
return torch.from_numpy(df).float()
X_traint = df_to_tensor(X_train)
y_traint = df_to_tensor(y_train)
X_testt = df_to_tensor(X_test)
y_testt = df_to_tensor(y_test)
# create pytorch dataset
train_ds = TensorDataset(X_traint, y_traint)
test_ds = TensorDataset(X_testt, y_testt)
# create data loaders
batch_size = 5
train_dl = DataLoader(train_ds, batch_size, shuffle=True)
test_dl = DataLoader(test_ds, batch_size, shuffle=False)
# model architecture
class BinaryNetwork(nn.Module):
def __init__(self, input_size, output_size):
super().__init__()
self.l1 = nn.Linear(input_size, 64)
self.l2 = nn.Linear(64, 32)
self.l3 = nn.Linear(32, 16)
self.out = nn.Linear(16, output_size)
def forward(self, x):
x = self.l1(x)
x = F.relu(x)
x = self.l2(x)
x = F.relu(x)
x = self.l3(x)
x = F.relu(x)
x = self.out(x)
return torch.sigmoid(x) # scaling values between 0 and 1
input_size = 13 # number of features
output_size = 1
model = BinaryNetwork(input_size, output_size)
loss_fn = nn.BCELoss() # Binary Cross Entropy
optim = torch.optim.Adam(model.parameters(), lr=1e-3)
model
BinaryNetwork(
(l1): Linear(in_features=13, out_features=64, bias=True)
(l2): Linear(in_features=64, out_features=32, bias=True)
(l3): Linear(in_features=32, out_features=16, bias=True)
(out): Linear(in_features=16, out_features=1, bias=True)
)
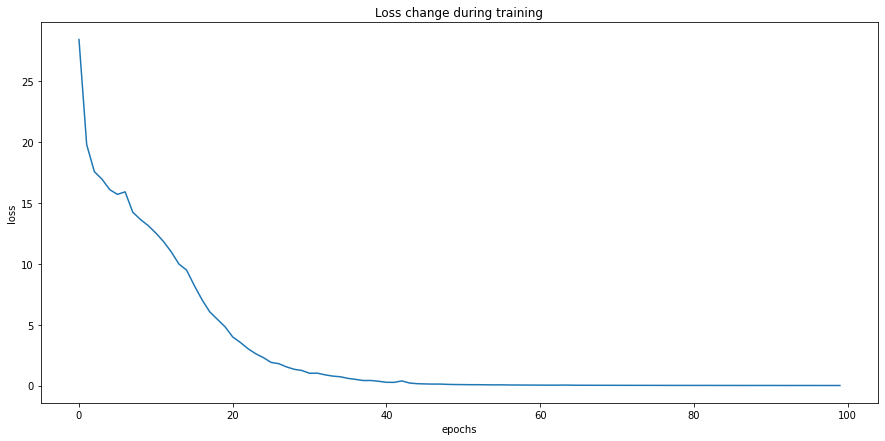
epochs = 100
losses = []
for i in range(epochs):
epoch_loss = 0
for feat, target in train_dl:
optim.zero_grad()
out = model(feat)
loss = loss_fn(out, target.unsqueeze(1))
epoch_loss += loss.item()
loss.backward()
optim.step()
losses.append(epoch_loss)
# print loss every 10
if i % 10 == 0:
print(f"Epoch: {i}/{epochs}, Loss = {loss:.5f}")
Epoch: 0/100, Loss = 0.79641
Epoch: 10/100, Loss = 0.03637
Epoch: 20/100, Loss = 0.07704
Epoch: 30/100, Loss = 0.02023
Epoch: 40/100, Loss = 0.00084
Epoch: 50/100, Loss = 0.00000
Epoch: 60/100, Loss = 0.00001
Epoch: 70/100, Loss = 0.00000
Epoch: 80/100, Loss = 0.00018
Epoch: 90/100, Loss = 0.00029
# plot losses
graph = sns.lineplot(x=[x for x in range(0, epochs)], y=losses)
graph.set(title="Loss change during training", xlabel='epochs', ylabel='loss')
plt.show()
# evaluate the model
y_pred_list = []
model.eval()
with torch.no_grad():
for X, y in test_dl:
y_test_pred = model(X)
y_pred_tag = torch.round(y_test_pred)
y_pred_list.append(y_pred_tag)
# convert predictions to a list of tensors with 1 dimention
y_pred_list = [a.squeeze() for a in y_pred_list]
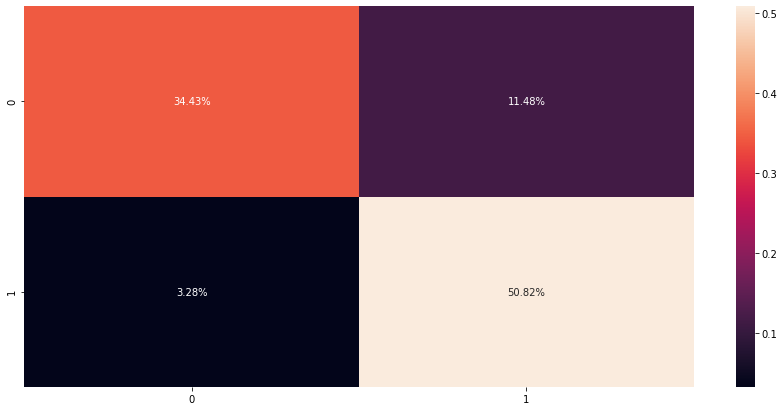
# check confusion matrix (hstack will merge all tensor lists into one list)
cfm = confusion_matrix(y_test, torch.hstack(y_pred_list))
sns.heatmap(cfm / np.sum(cfm), annot=True, fmt='.2%')
<AxesSubplot:>
# print metrics
print(classification_report(y_test, torch.hstack(y_pred_list)))
precision recall f1-score support
0 0.91 0.75 0.82 28
1 0.82 0.94 0.87 33
accuracy 0.85 61
macro avg 0.86 0.84 0.85 61
weighted avg 0.86 0.85 0.85 61

A deep dive into the intricacies of deploying custom models to Amazon SageMaker
How to create a new novel datasets from a few set of images.

Data Science Project

Data Science Project